Project description
Analysis Goals
The goals of this example case-study are to train the user on:
- Downloading files from the ImmPort Data Repository directly to the Analysis Workspace platform.
- Performing an analysis using Tools which are available on the Platform. This project is based on analysis performed in the paper MetaCyto: A tool for automated meta-analysis of mass and flow cytometry data (Hu, et al., 2018).
Analysis Context
In this study, the researchers developed the MetaCyto R package to enable automated meta-analysis of cytometry datasets, including data from both conventional flow cytometry and CyTOF data. These data are normally incomparable due to different methods and model parameters, but the advent of MetaCyto makes this possible.
MetaCyto is able to accurately identify common cell populations across studies without parameter tuning requirements or tedious user intervention; it unifies the readings from the different cytometry methods into a nom-metric space. It then applies hierarchical models to robustly estimate the effects of factors of interest (e.g. age, ethnicity, or vaccination) on the cell populations using data across all selected input studies and cytometry data types.
To test the utility of MetaCyto, the authors performed a joint analysis of 10 human immunology cytometry datasets contributed by four different institutions (Ledgerwood, et al., 2012; Obermoser, et al., 2013; Wertheimer et al., 2014; Whiting, et al., 2015).
Altogether, this analysis spanned 2926 whole blood or peripheral blood mononuclear cells (PBMC) samples from 984 healthy subjects, which were acquired using either flow cytometry or CyTOF with a diverse set of markers. For this MetaCyto Public Project samples from two studies, Study ID SDY312 and SDY420, will be used to recreate the analysis from Hu, et al. (2018).
Analysis Results
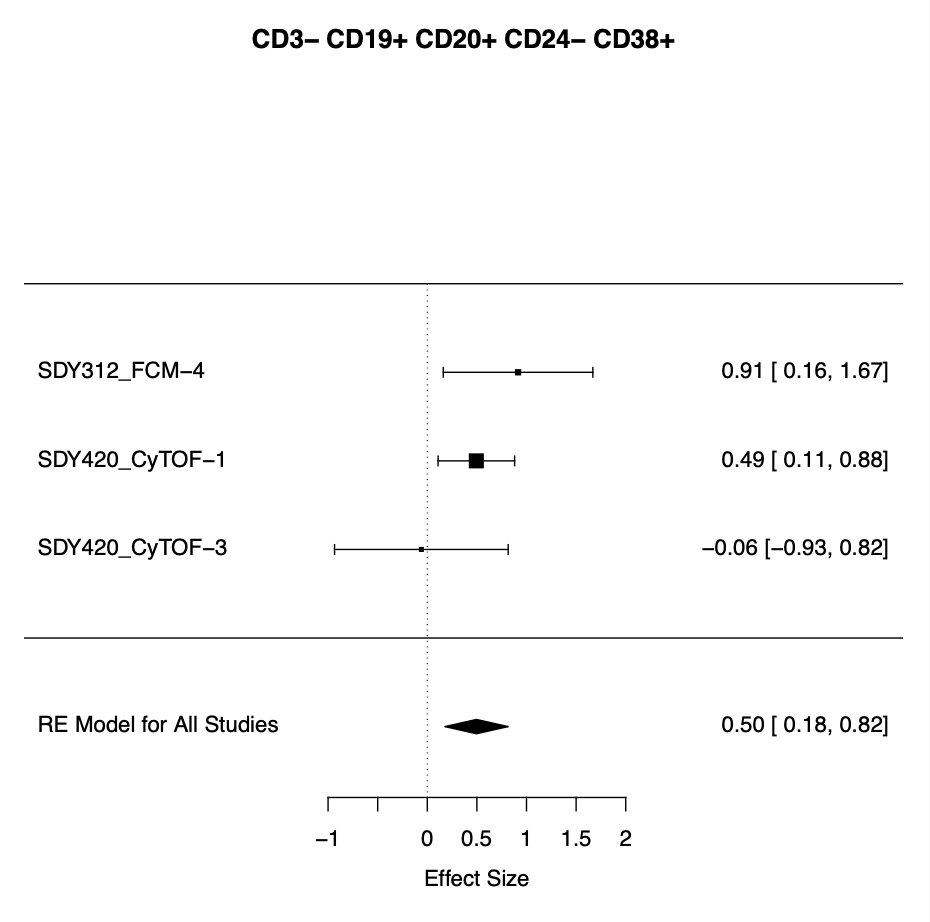
By jointly analyzing 2 out of 10 datasets used in the MetaCyto publication and comparing the results, the user will have replicated a result from the publication: The authors provided significant results showing increased naive B cell proportions in the Asian subject population compared to Caucasian subject population.
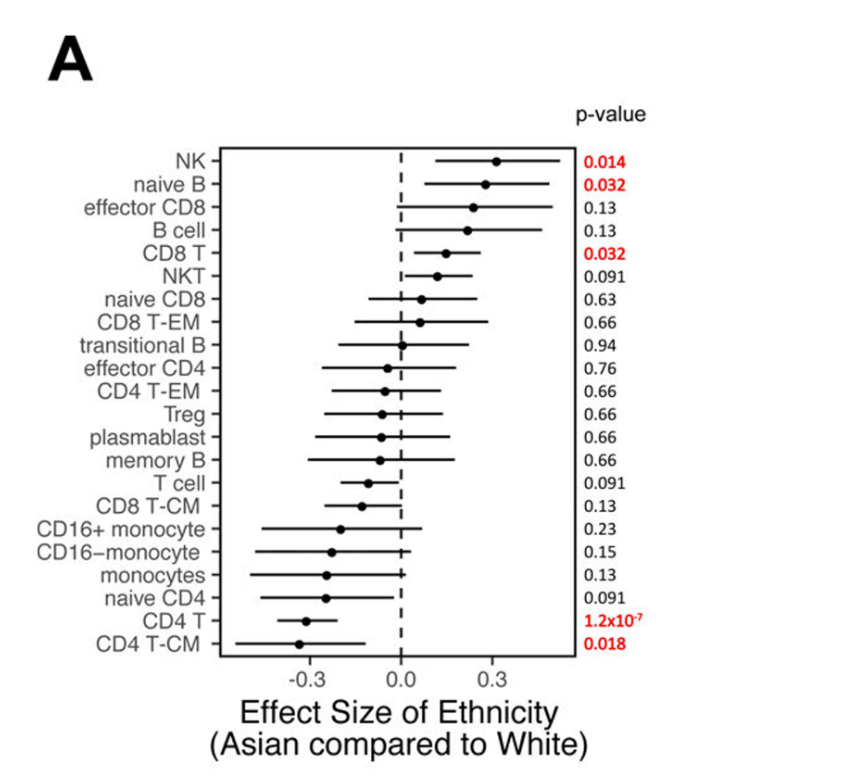
(Above).Figure 5a from Hu et al. (2018) showing proportions of different cell types in Asian populations (horizontal bars and circles) to Caucasian populations (vertical dashed line).
(Below). Results produced from the MetaCyto Public Project, replicating the results for one of these cell types (naive B cells).
Updated 3 months ago
