CCLE data
Seven Bridges is committed to providing CAVATICA users with the most up-to-date version of the CCLE dataset that is available from the NCI Genomic Data Commons (GDC). In keeping with this commitment, CAVATICA transitioned from hosting the CGHub version of this dataset to the GDC Legacy Archive Data Release 11.0 version on July 10, 2018. As of this date, all files accessible via the Data Browser and the API correspond to files in the GDC Legacy Archive. Files that were added to individual projects before this date and are no longer represented in the new dataset version will no longer be accessible via those projects but may be obtainable from the GDC archive by contacting the GDC Help Desk. We look forward to continuing to collaborate with the GDC in the months ahead to ensure the timely availability through CAVATICA of new data releases for this dataset.
The Cancer Cell Line Encyclopedia (CCLE) public project contains large Open Access files from the CCLE which you can use on CAVATICA.
On this page:
Overview
The CCLE is made possible through a collaboration between the Broad Institute, the Novartis Institutes for Biomedical Research, and the Genomics Institute of the Novartis Research Foundation to perform detailed genetic and pharmacologic characterization of a large number of human cancer models.
The CCLE public project contains Open Access sequencing data (in the form of reads aligned to the hg19 broad variant reference genome) for nearly 1000 cancer cell line samples, as available from cgHub on May 11, 2016.
Open AccessYou don't need special access or authorization status to use the data in this project. In fact, any data you copy from this public project into your own projects will not count towards your storage. This data is ideal for interrogating the genomic landscape of cancer cell lines, testing new analysis methods, or getting to know CAVATICA.
What's contained in the project?
The CCLE public project contains the following distribution of samples and files by experimental strategy.
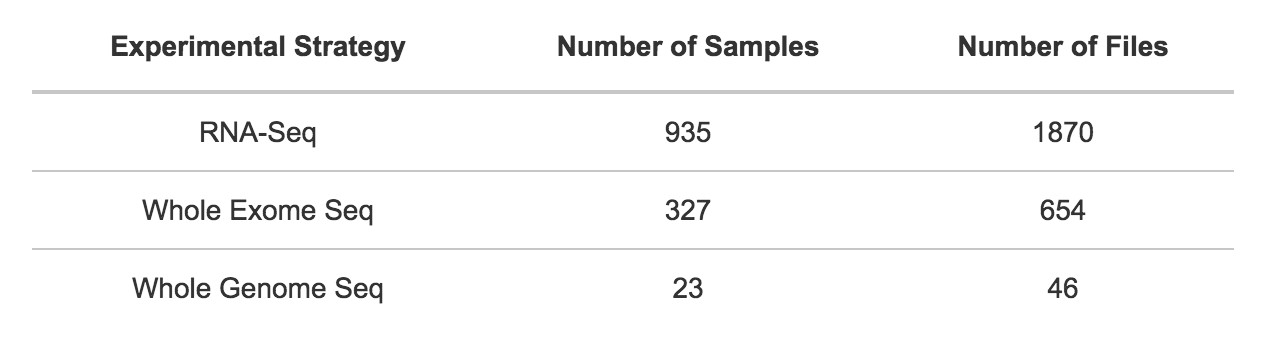
Additional array data and processed data are available from the CCLE data portal.
Access the CCLE public project
To access the CCLE public project:
- Click on Public projects from the top navigation bar.
- Select Cancer Cell Line Encyclopedia (CCLE), as shown below.
You'll be taken to the main dashboard of the CCLE public project.
Use the CCLE public project
All CAVATICA users automatically have copy permissions for this project. This means that while you while you cannot upload data or tools to the project or execute any of the workflows contained within it, you can copy the available data, tools, and workflows to your own projects on CAVATICA and execute analyses in your own projects.
You have the options to:
- Use the entire project - Copy the entire project to replicate analyses or conduct further analysis on CCLE data.
- Select and use a subset of the data in your own project - Copy the selected data to use within your own analyses.
Use the entire project
To help you get started, the Seven Bridges bioinformatics team has run some tasks on the CCLE data using Seven Bridges public apps and reference files.
While you cannot directly execute the workflows in the CCLE public project, you can make a copy of the entire project to perform further analyses.
Copy the entire project
To copy the entire project:
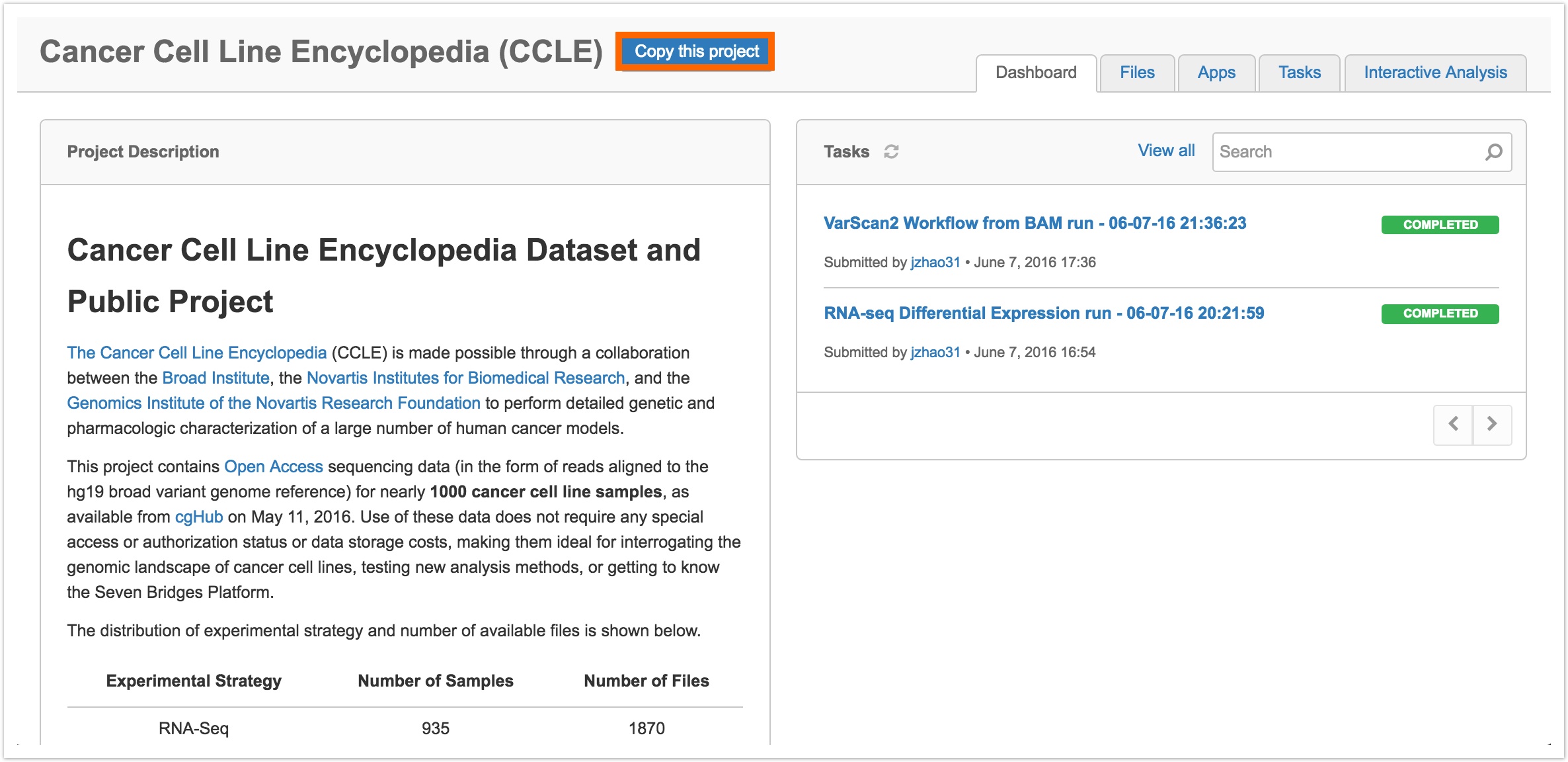
- Access the CCLE public project by selecting Cancer Cell Line Encyclopedia (CCLE) from Public projects in the top navigation bar.
- Click Copy this project, next to the project's title, as shown below.
- In the pop-up window, you can name your copy of the project and select a billing group.
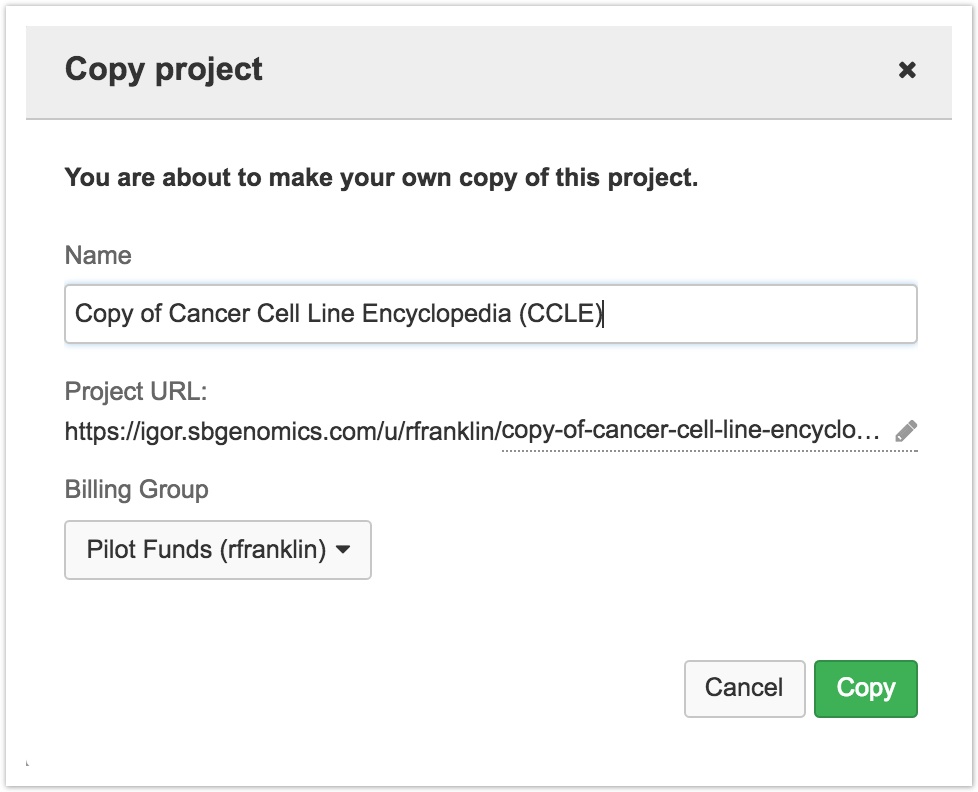
- Once you've customized the details, click Copy to copy the entire project.
You'll be redirected to the dashboard of your cloned project when it is ready, as shown below.
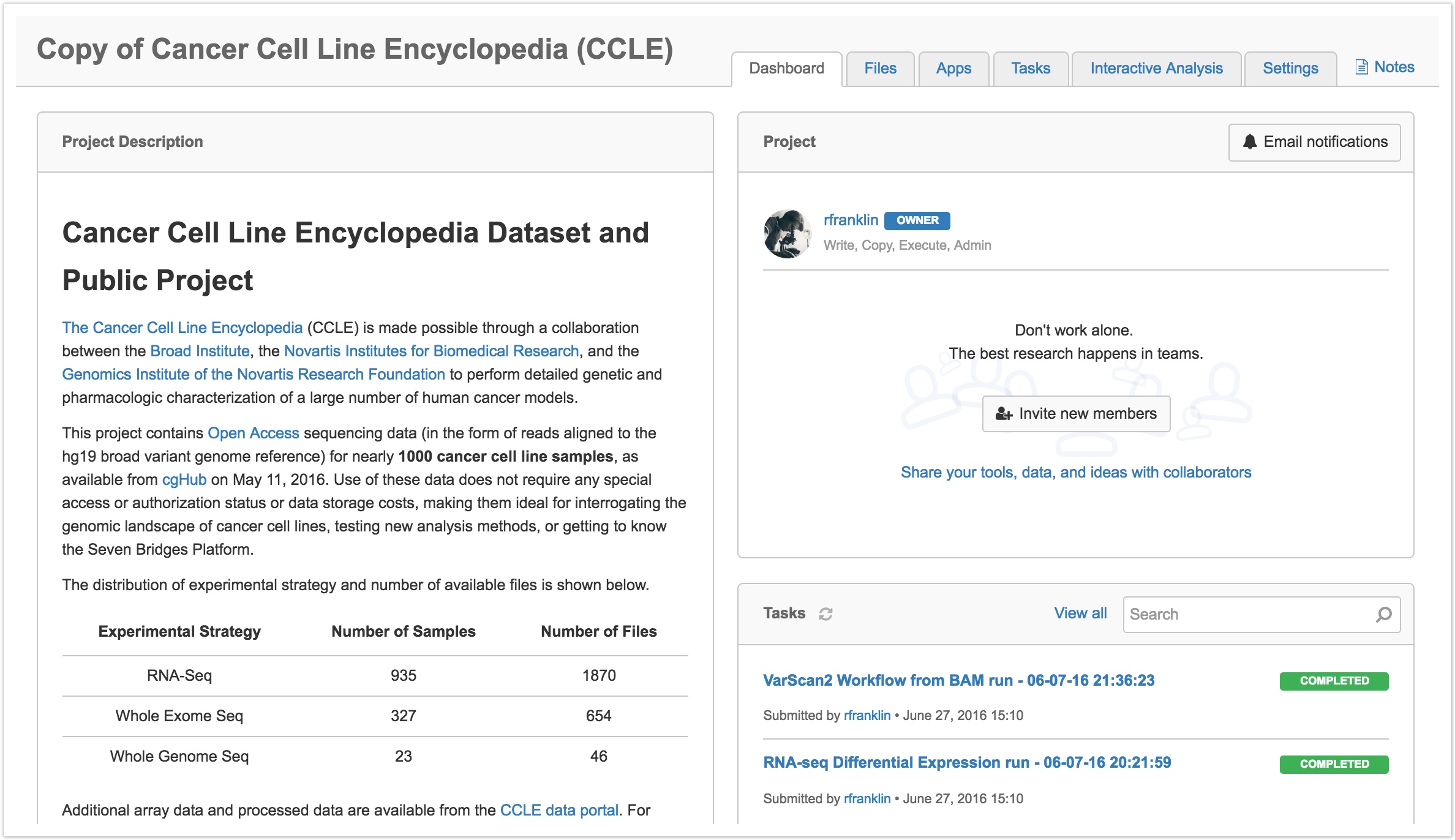
You can now use the data and workflows contained within this project to conduct your own analyses.
Run a task in a copied CCLE project
For instance, you can replicate one of the tasks in the project. The RNA-seq Differential Expression task in the copied project is a great starting point to run other differential expression analyses. All you have to do is change the input files.
Once you've copied the project, you can run tasks in it.
- Navigate to the Tasks tab on your copied CCLE project's dashboard.
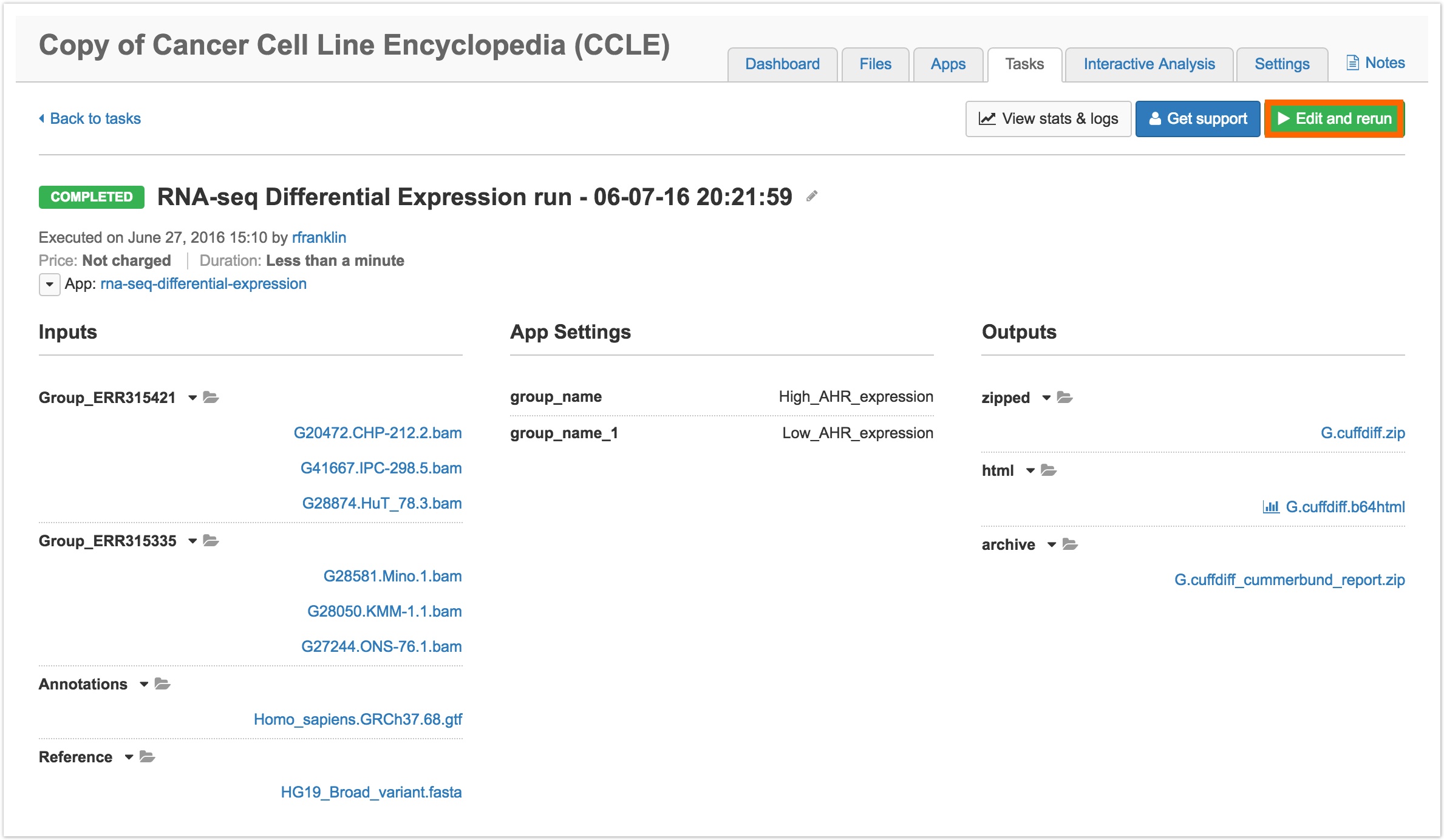
- Select one of the previously run tasks.
- Click Edit and rerun in the upper right hand corner, as shown below.
- You'll be redirected to the DRAFT task page, where you can select different inputs. Simply click Pick file(s), as you can see in the image below, to select different files.
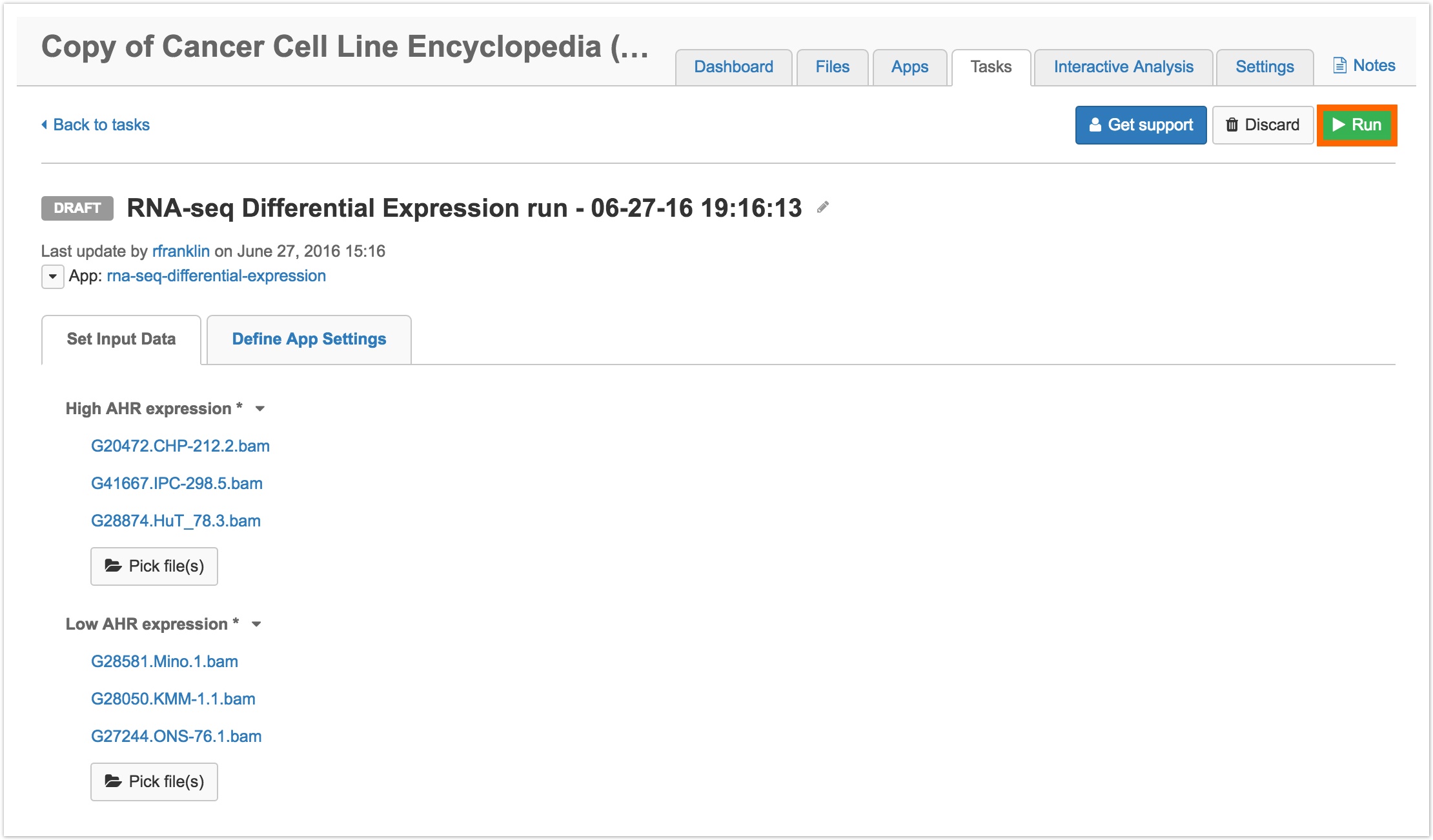
- When you've selected your inputs, click Run in the upper righthand corner, as shown above.
Use a subset of the data
Instead of cloning the entire project, you can choose to select and copy a subset of the data.
To copy a subset of the data:
- Access the CCLE public project by selecting Cancer Cell Line Encyclopedia (CCLE) from Public projects in the top navigation bar. You'll be taken to the project dashboard of the CCLE public project, as shown below.
.jpeg)
- Click on the Files tab in the upper righthand corner. This will take you to the Files page for the CCLE project, as shown below.
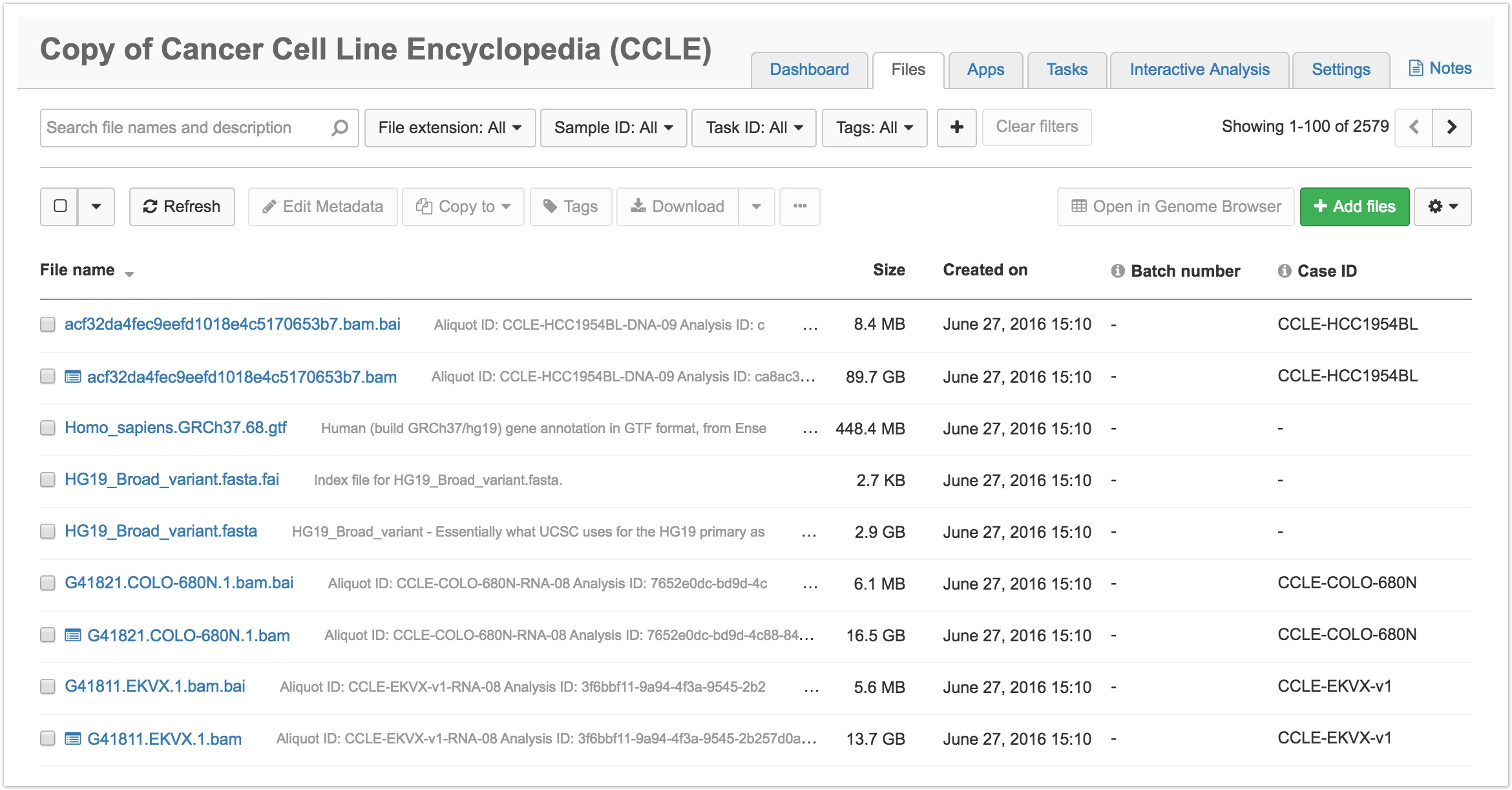
- Filter or search for the desired files. You can filter by:
- Keywords - You can use the search bar at the top of the page to find files by entering the file name or notes associated with a file.
- Metadata fields - Next to the search bar, you will see drop-down menus for the metadata fields File extension, Sample ID, and Task. Selecting a particular metadata value from one of these menus displays only files that match the value. You can add additional drop-down menus to filter by other metadata fields by clicking the + icon.
- You can choose specific files by selecting the corresponding checkbox in front of the file name.
- Select as many files as you desire and click Copy to.
- Select your desired project from the drop-down menu.
Now, you can start using the CCLE files you've added to your personal project in your own analysis.
Updated 4 months ago
