MetaCyto Analysis Walkthrough
This page contains detailed, step-by-step instructions to follow while carrying out the analysis through the Meta-Analysis Of Cytometry Data From ImmPort Data Repository (MetaCyto) Public Project. This example project collects two different cytometry methodologies, and makes them comparable (Flow Cytometry and CyTOF). For this study, we compare data from two different studies of immune cells in populations of different ethnicity, Caucasian and Asian.
Step 1: Register for an account
- Access CAVATICA.
- Click Create an account and fill out the form.
- You will receive an email from Seven Bridges to confirm your email. Once you confirm your email, you can log into CAVATICA.
Step 2: Request pilot funds
Every user on the Platform has to be assigned to a billing group. To get you started, you can apply for pilot funds, which will be added to a ‘Pilot Funds’ billing group.
Once your account has been created, send an email to [email protected] and request that pilot funds are added to your Pilot Funds billing group.
Please make sure you get access to the pilot funds before moving to the STEP 3.
Step 3: Copy and Run the Meta-analysis of Cytometry Data From ImmPort Data Repository Public Project
To perform the Meta-analysis, you will first copy the Public Project, use the Data Studios to download data, and use Tools and Workflows to run the analyses.
Public Projects contain all necessary files, instructions, and Tools necessary to perform the specified analysis(es).
In order to perform the analysis yourself, you will need to make your own copy as the Public Project serves as a repository for all users, and as such, is not editable.
Copy the public project
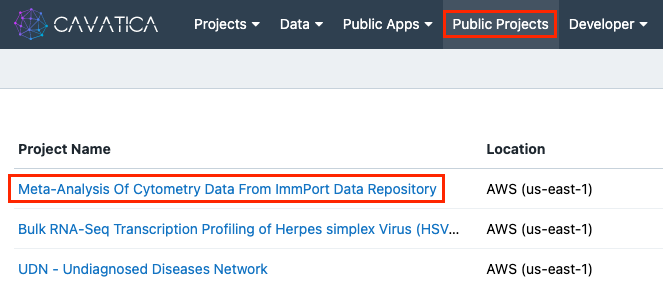
- Click Public Projects on the top navigation bar.
- Click "Meta-Analysis Of Cytometry Data From ImmPort Data Repository".
-
Click the ‘i’ icon next to the public project title, and then click Copy project. The ‘Copy Project’ window will appear.
-
Under "Billing group", select your pilot funds group (to which pilot fund credits were awarded in STEP 2).
-
Click Copy. The project is copied and the project dashboard is displayed.
The next step is initializing Data Studio for SDY312. Read below.
Initialize Data Studio for SDY312
- Click on Data Studio in the Project toolbar. You will observe identical Data Studio Tasks, except for the study number. We will start with “”.
- Click Start next to "Cytometry Data Download From SDY312".
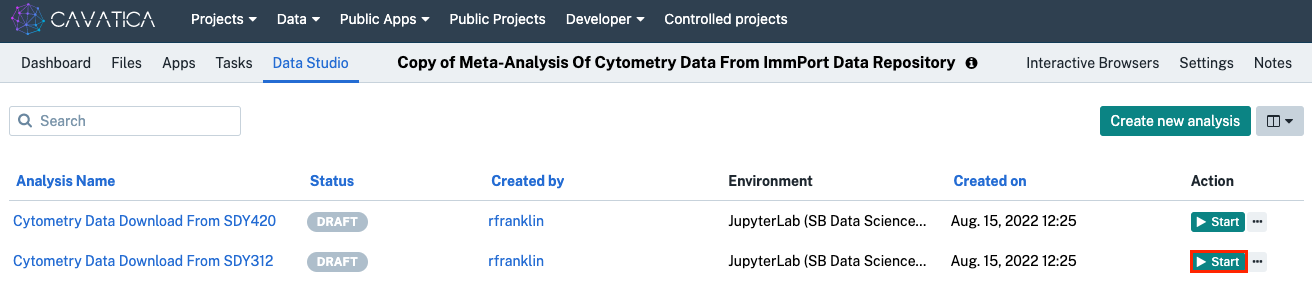
If the initialization halts at any of the three steps for more than 5 minutes, click Stop, wait for the Data Studio to finish closing, and reopen.
Once the initialization is successful, the Jypyter notebook environment is displayed.
Enter your credentials
To successfully download your study data, you will need to enter your ImmPort Data Repository account credentials.
- Double click the red and white box item on the left, it will open a new tab to the right.
- Scroll down to the "User Inputs" section and change the Study ID section to "study_id=312"

- Locate the
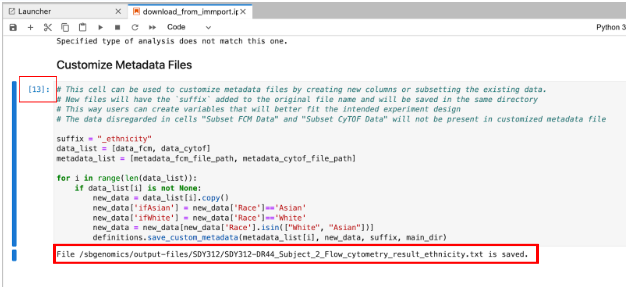
user_nameandpasswordparameters and populate them with your credentials. - Locate the
type_of_analysisparameter and enter "FCM" between the double quotations. - Locate the
metadata_fcm_filepathparameter and enter "/SDY312/SDY312-DR47_Subject_2_Flow_cytometry_result.txt"
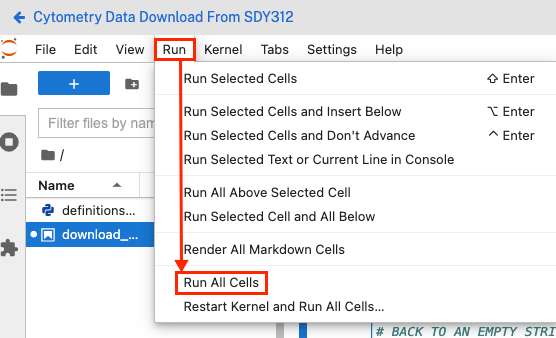
Run the analysis
To run the analysis after setting your credentials:
- Open the Run menu.
- Click Run All Cells.
Confirm successful execution and export data
Navigate towards the bottom, and if you see a list populating in real time, as shown below, your analysis is performing as expected.
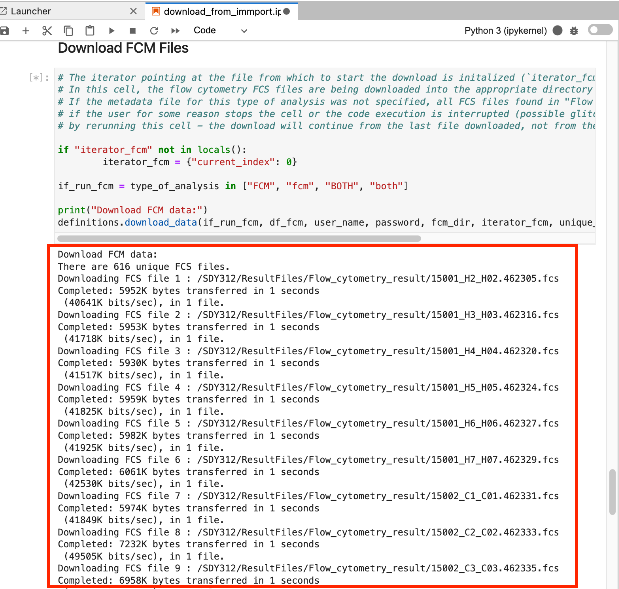
[Square brackets] to the right number the steps in the code, and the current executing step will change to a “[*]”.
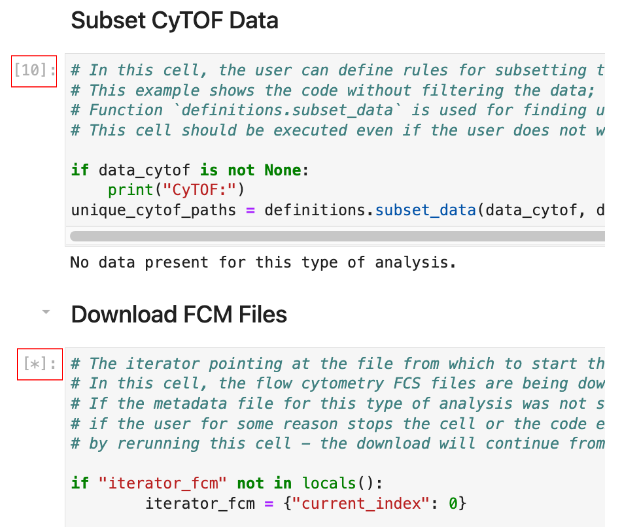
You can navigate away from this window but DO NOT click stop OR close the window until your download from ImmPort is complete.
SDY312 data download will take approx. 1 hour. It will look like the screen below when complete.
Click Stop. The Window will close and you will be returned to the Project Dashboard.

Repeat to download data from ImmPort for SDY420
After successful completion of data collection for SDY312, you will perform the same procedure for SDY420 (remember to change the study_id parameter), but instead download CyTOF data for this study, as opposed to the previous Flow Cytometry data. Steps 4&5 for SDY420 are modified as follows:
4. Locate the type_of_analysis parameter and enter "CyTOF" between the double quotations.
5. Locate the metadata_fcm_filepath parameter and enter "/SDY420/SDY420-DR47_Subject_2_CyTOF_result.txt"
SDY420 data download will take approx. 25 mins.
Confirm successful download of data
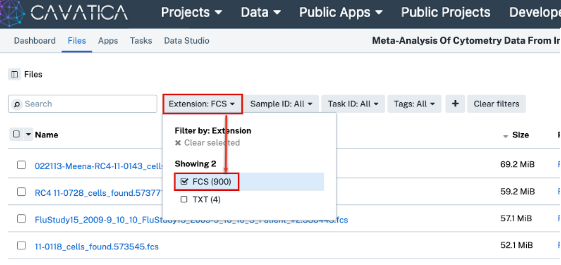
Click the Files tab in the top toolbar, and confirm that there are 900 “.FCS” files present in your project.
Step 4: Run the ‘MetaCyto Get Markers’ Tool to retrieve metadata used as variables in your meta-analysis
This preprocessing step retrieves the names for different cell types across your studies.
- Click Apps in the project toolbar.
- Click Run for the ‘MetaCyto Get Markers’ tool. A new window will open.
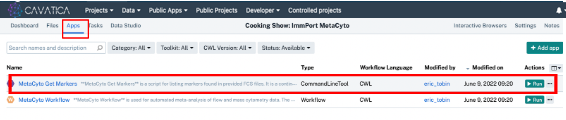
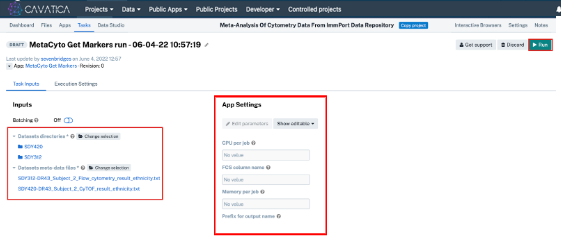
- Click “Select file(s)” next to “Database Directories”. Select studies “SDY420” and “SDY312”.
- Click “Select file(s)” next to “Database meta-data files”. Select “SDY312-DR43_Subject_2_Flow_cytometry_result_ethnicity.txt” and “SDY420-DR43_Subject_2_CyTOF_result_ethnicity.txt”.
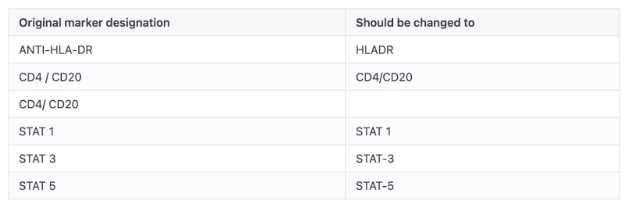
Studies may have a slightly different way of labeling identical cell types, which will need harmonization. For this study, there are several data discrepancies, as displayed below. When conducting your own research, you will need to create a file similar to the one provided (‘new_markers_312_420.txt’) in order to standardize your data.
Run the analysis
After you have retrieved your meta-data, you will run the meta-analysis.
- Click Tasks in the project toolbar.
- Click Run for the ‘MetaCyto Workflow run - 02-02-23 15:53:53’ tool. A new window will open for a task that is prepopulated with parameters; you will add your data into this prebuilt execution of the App.
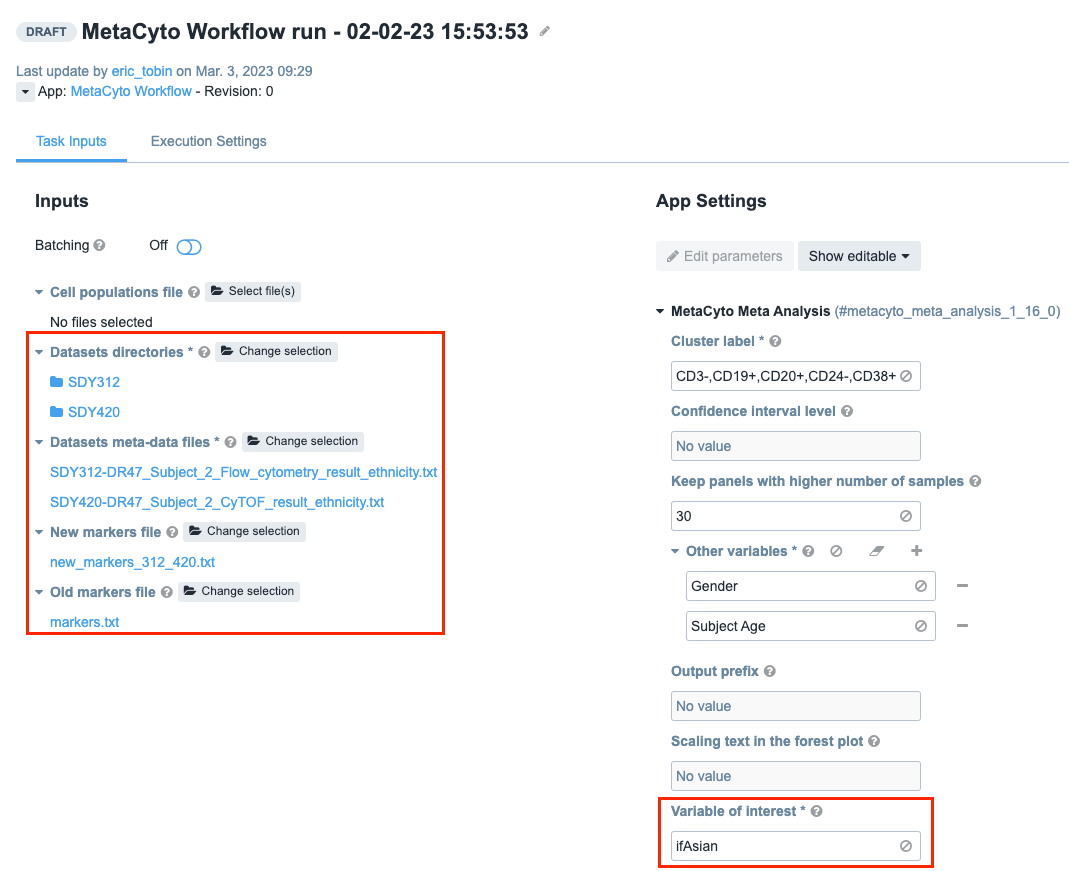
- Click Select file(s) next to “Dataset directories”. Select “SDY420” and “SDY312”.
- Click Select file(s) next to “Datasets meta-data files”. Select “SDY312-DR43_Subject_2_Flow_cytometry_result_ethnicity.txt” and “SDY420-DR43_Subject_2_CyTOF_result_ethnicity.txt”.
- Click Select file(s) next to “New markers file”. Select “new_markers_312_420.txt”.
- Click Select file(s) next to “Old Markers File”. Select the file you created from the previous Task: “markers.txt”.
App Settings - Examine the variable of interest parameter(case sensitive): ifAsian.
- Click Run in the upper part of the page. This will automatically save and export your results when complete (~10 mins).
Analyze and interpret results
The source paper by Hu et al. (2018) contain comparable results, except using more studies and analyzing more cell types.
Your results will match those shown below:
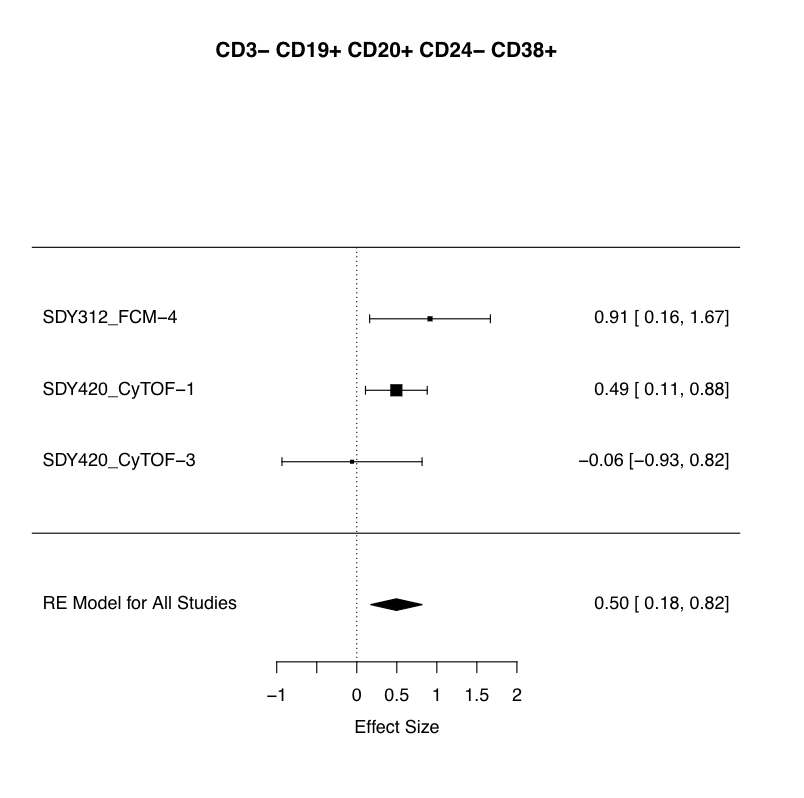
This figure and associated statistical results provide evidence for overrepresentation of B-cells in Asian subjects (box and lines; diamond) as compared to the null population (dashed line) of Caucasian subjects.
Updated 3 months ago
