Introduction to datasets
On this page:
CAVATICA hosts both The Cancer Genome Atlas (TCGA) and the Cancer Cell Line Encyclopedia (CCLE), two datasets you can use in your genomics analyses. On this page, learn more about these datasets as well as their underlying metadata structure.
The following public datasets are made available on CAVATICA. Note that these public datasets are different from CAVATICA datasets.
The Cancer Genome Atlas (TCGA)
TCGA is one of the world’s largest cancer genomics data collections, including more than eleven thousand patients, representing 33 cancers, and over half a million total files. Data collection for TCGA began in 2006 as a joint effort by the National Cancer Institute (NCI), National Human Genome Research Institute (NHGRI), the National Institutes of Health (NIH), and the U.S. Department of Health and Human Services. CAVATICA provides powerful methods to query and reproducibly analyze TCGA data by itself or in conjunction with your own data.
TCGA data is made available on CAVATICA through an integration with the Seven Bridges Cancer Genomics Cloud (CGC). TCGA on CAVATICA includes both Open and Controlled Data. While all data in TCGA is stripped of direct identifiers, DNA information is inherently unique to an individual. Two types of data access ‘tiers’ (Open Data and Controlled Data) have been put in place to balance the desire to make TCGA data as widely available as possible while ensuring that the rights of study participants are well protected. You can access TCGA Open Data on CAVATICA after you are authenticated and agree to data use policies. In addition, you can obtain access to Controlled Data through the NIH via the Database of Genotypes and Phenotypes (dBGaP) site.
Learn more about TCGA Data on CAVATICA, permissions required to access TCGA data, and the TCGA metadata schema.
###Cancer Cell Line Encyclopedia (CCLE)
The Cancer Cell Line Encyclopedia (CCLE) is a project performing detailed genetic and pharmacologic characterization of a large number of human cancer cell lines, permanently established cell cultures derived from patients that will proliferate indefinitely given appropriate fresh medium and space. The CCLE is the result of a collaboration between the Broad Institute, the Novartis Institutes for Biomedical Research, and the Genomics Institute of the Novartis Research Foundation.
CCLE contains Open Access sequencing data (in the form of reads aligned to the hg19 reference genome) for nearly 1000 cancer cell line samples. CAVATICA hosts the CCLE dataset in the form of a read-only public project which contains cell line samples as available from cgHub on May 11, 2016. You have automatic access to all CCLE data on CAVATICA.
Learn more about the CCLE public project and the CCLE metadata schema.
GDC Datasets Update Policy
Seven Bridges is committed to providing CAVATICA users with up-to-date versions of the datasets that are available from the NCI Genomic Data Commons (GDC). Therefore, we have a clearly formulated set of rules that apply to updates of GDC datasets that are available through CAVATICA:
- We aim to update the data on CAVATICA within 30 day of release by the GDC.
- The time frame for alignment of datasets available through CAVATICA with the current GDC data release is within 30 days of the release by GDC.
- If a GDC data release includes redaction of files from a dataset, the affected files will be available on CAVATICA for an additional 30 days. After that, you will need to contact the GDC for information on how to retain access to redacted files.
- Re-running queries executed in the past may return slightly different results due to updates in the datasets from the GDC. This is expected as datasets are dynamic and version updates can introduce file updates or redactions, and queries will return the most up to date version of files. This applies both to the queries made through the Data Browser and through the Datasets API.
##Metadata for public datasets on CAVATICA
Metadata is data about the genomic information carried by files. It is data about the time, place, and manner in which the genomic data was obtained as well as the genomic data's source and type. You can use metadata on CAVATICA to browse and query datasets. Metadata describing datasets on CAVATICA consist of properties which describe the entities of each dataset.
Entities are particular resources with UUIDs, such as files, cases, samples, and cell lines. These can be the subject of your query.
Properties can either describe an entity or relate that entity to another entity. For instance, properties include an entity's vital status, gender, data format, or experimental strategy.
View the metadata schema, which includes a list of entities and their related properties, for the following datasets:
Below, learn how to start working with datasets via their metadata on the visual interface.
##Explore datasets using the visual interface
The The Data Browser allows you to explore datasets using an interactive graphical interface. Start by building queries to filter data using various metadata attributes. Then, access these files for further analysis.
To access the Data Browser, click on Data on the top navigation bar and select Data Browser. You'll see the screen below. Here, you can select the dataset to query.
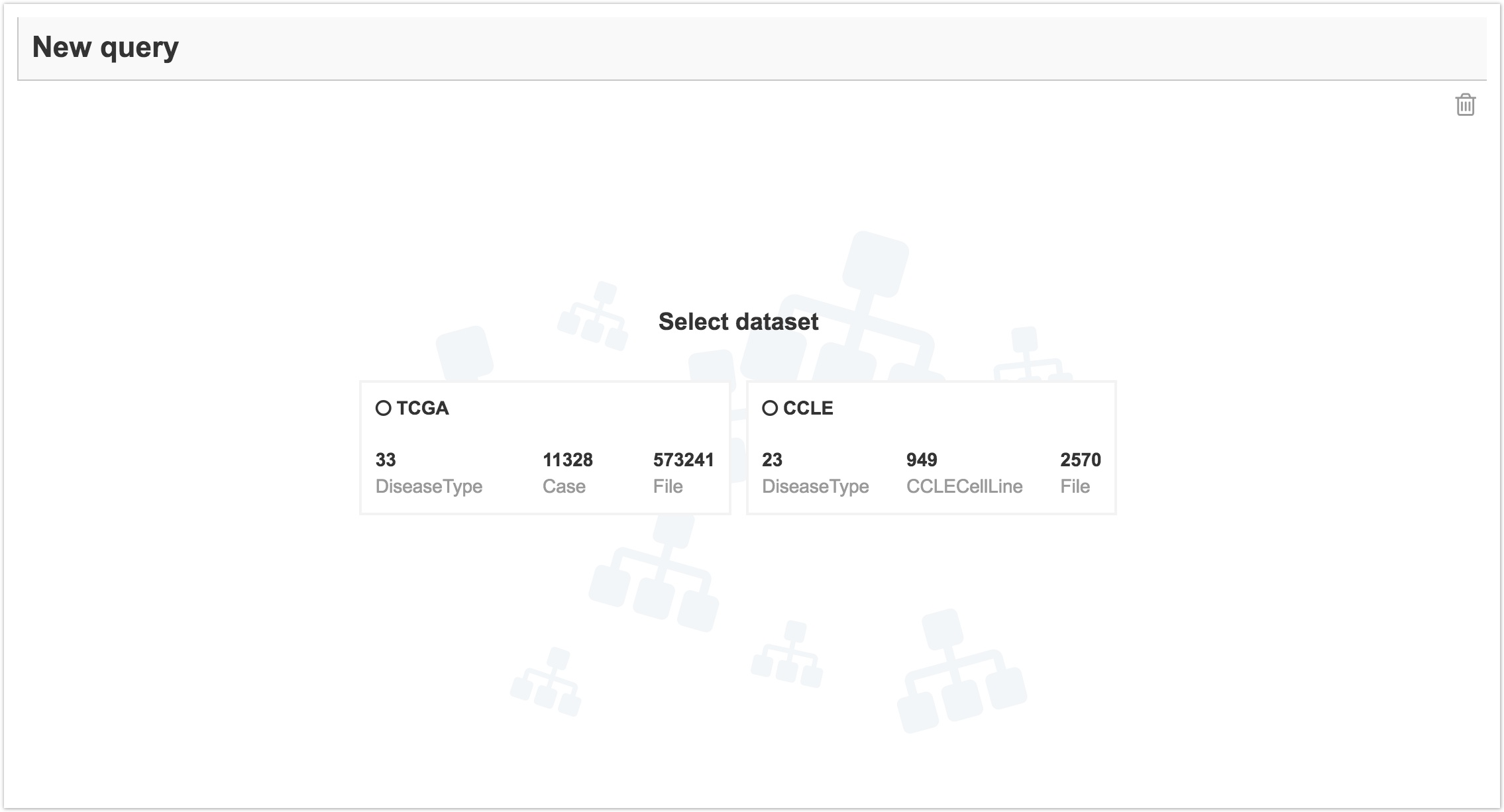
Take advantage of pre-built example queries or build your own from scratch using metadata entities and properties. Learn more about queries in the Data Browser.
Once you've located specific files using a Data Browser query, you can access this data for further analysis.
TCGA Controlled DataRemember, TCGA contains both Open and Controlled Data. When you attempt to access TCGA data from CAVATICA, you will be asked to authenticate with dbGaP. You will only be able to access the data for which you are approved. Learn more about TCGA data access on CAVATICA.
Related pages
Updated 2 months ago
